Biological Sciences | Simulation
Identifying Reaction Coordinates for Complex Protein Dynamics
Proteins play an essential role in nearly all biological processes. By investigating proteins and their functions, scientists are providing insights to drive drug development, further our understanding of disease mechanisms, and advance many other areas of biomedical research. With help from ALCF supercomputers, a team from University of Illinois Chicago (UIC) has made an important breakthrough in understanding how proteins function.
Challenge
The primary goal of protein science is to understand how proteins function, which requires understanding the dynamics responsible for transitions between different functional structures of a protein. If the exact reaction coordinates (the small number of essential coordinates that control functional dynamics) were known, researchers could determine the transition rate for any protein configuration and thoroughly understand its mechanism. Despite intensive efforts, identifying the exact reaction coordinates in complex molecules remains a formidable challenge.
Approach
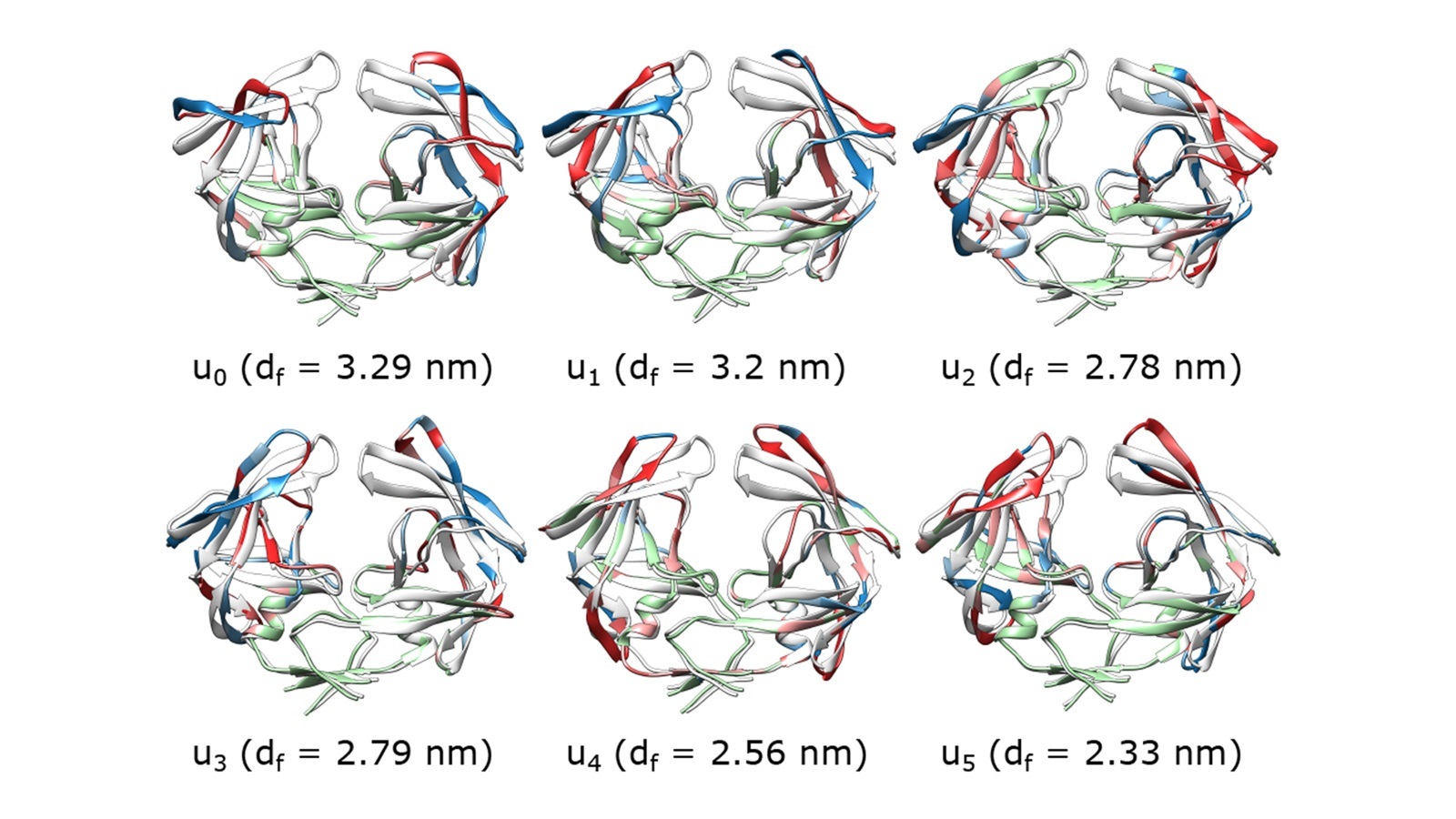
The UIC team employed their generalized work functional (GWF) method to study the flap opening process of HIV-1 protease, a complex protein and major drug target for combatting the HIV virus. GWF is a fundamental mechanical quantity rooted in Newton’s law. Using the transition path sampling method, the researchers leveraged the ALCF’s Theta supercomputer to generate 2,000 reactive trajectories that start from structures of HIV-1 protease with flaps in the semi-open state and end at structures with flaps in the open state. This data served as the input to the GWF method, which was used to pinpoint the exact reaction coordinates and determine the molecular mechanism of the flap opening process.
Results
As detailed in their paper in the Proceedings of the National Academy of Sciences, the team was able to identify the exact reaction coordinates for a major conformational change of a large functional protein for the first time. Their results show that the flap opening of HIV-1 protease has six reaction coordinates, providing the precise definition of collectivity and cooperativity in the functional dynamics of a protein. Success in determining the reaction coordinates enabled acceleration of this important process by 103 to 104 folds compared to regular molecular dynamics simulations. The team’s work demonstrates that the GWF method could potentially be applied to other problems in protein research, such as folding, entropic barriers, and reaction rates.
Impact
By successfully identifying the exact reaction coordinates for a complex protein for the first time, the team has made an important breakthrough toward understanding protein functional dynamics. Their work has far-reaching implications for both biomedical research and protein engineering, providing insights that are crucial for designing drugs, fighting drug resistance, and developing artificial enzymes that can complete desired functions.