|
High-Performance Computational Chemistry: NWChemEx
As the original NWChem code—an ab initio computational chemistry software package that includes quantum chemical and molecular dynamics functionality—is nearly a quarter-century old, the NWChemEx developers decided update the application by rewriting from the ground up, with the ultimate goal of providing the framework for next-generation molecular modeling. The new, exascale-ready code is capable of enabling chemistry research on a variety of leading-edge computing systems.
Challenge
The NWChemEx developers aim to restructure core functionality—including the elimination of longstanding bottlenecks associated with the generally successful NWChem code—concurrent with the production of sophisticated physics models intended to leverage the computing power promised by the exascale era. As one component of this strategy, the developers have adopted the Aurora-supported DPC++ programming model as one of its development platforms.
From a design point-of-view, the development team gives equal weight and consideration to physics models, architecture, and software structure, in order to fully harness large-scale HPC systems. To this end, NWChemEx incorporates numerous modern software-engineering techniques for C++, while GPU compatibility and support have been planned since the project’s initial stages, thereby orienting the code to the demands of exascale as matter of constitution.
In order to overcome prior communication-related bottlenecks, the developers have localized communication to the greatest possible extent.
To help localize communication and thereby reduce related bottlenecks, NWChemEx is being geared such that CPUs handle communication protocols as well as any other non-intensive components (that is conditional-structure-based algorithms). Anything else—anything “embarrassingly parallel” or computationally expensive—is to be processed by GPU.
For Intel hardware, the developers employ Intel’s DPC++ Compatibility Tool to port any existing optimized CUDA code and translate it to DPC++. The Compatibility Tool is sophisticated enough that it reliably determines apposite syntax in translating abstractions from CUDA to SYCL, greatly reducing the developers’ burden. Subsequent to translation, the developers finetune the DPC++ code to remove any redundancies, inelegancies, or performance issues introduced by automation.
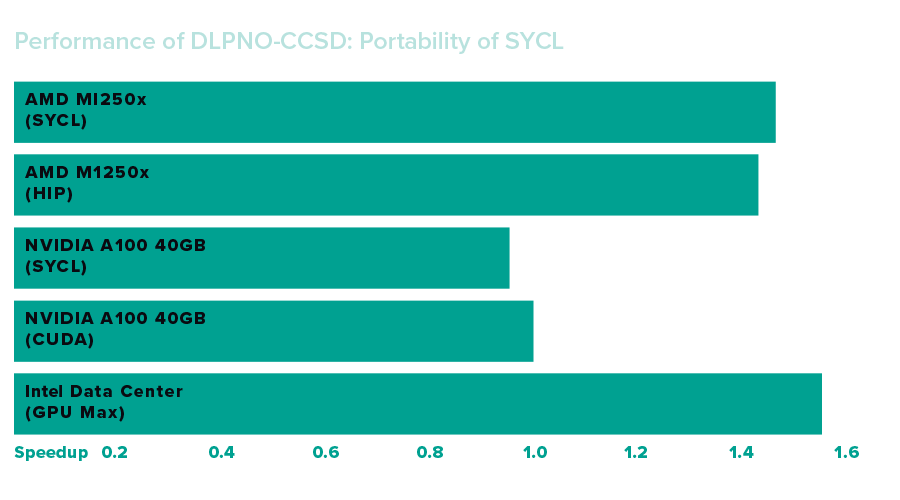
Performance Results
The NWChemEx simulations were carried out in 2023 for both single-GPU performance evaluations and large-scale demonstration runs involving up to 512 nodes. Canonical coupled cluster singles and doubles (CCSD) methods for molecular description showed faster performance on Aurora than was achieved using previous-generation systems, while the domain-based local pair natural orbital coupled-cluster method with single-, double- and perturbative triple excitations (DLPNO-CCSD) gave approximately the same performance compared to another tested system.
Impact
The NWChemEx project, when realized, has the potential to accelerate the development of next-generation batteries, drive the design of new functional materials, and advance the simulation of combustive chemical processes, in addition to addressing a wealth of other pressing challenges at the forefront of molecular modeling, including the development of stress-resistant biomass feedstock and the development of energy-efficient catalytic processes to convert biomass-derived materials into biofuels.